Background
The coronavirus disease 2019 (COVID-19) pandemic, caused by the SARS-CoV-2 pathogen, has burdened the healthcare and socioeconomic structures of many countries globally. To curb the viral transmission, several vaccines have been made in record time, and some of these vaccines are already rolling out in many pandemic-affected countries.
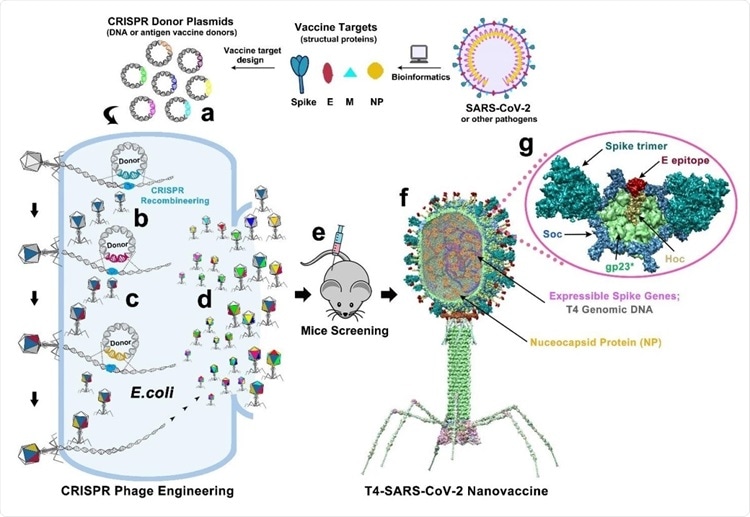
Design of T4-SARS-CoV-2 nanovaccine by CRISPR engineering. Engineered DNAs corresponding to various components of SARS-CoV-2 virion are incorporated into bacteriophage T4 genome. Each DNA was introduced into E. coli as a donor plasmid (a), recombined into injected phage genome through CRISPR-targeted genome editing (b). Different combinations of CoV-2 inserts were then generated by simple phage infections and identifying the recombinant phages in the progeny (c). For example, recombinant phage containing CoV-2 insert #1 (dark blue) can be used to infect CRISPR E. coli containing Co-V2 insert containing donor plasmid #2 (dark red). The progeny plaques obtained will contain recombinant phage #3 with both inserts #1 and #2 (dark blue plus dark red) in the same genome. This process was repeated to rapidly construct a pipeline of multiplex T4-SARS-CoV-2 vaccine phages (d). Selected vaccine candidates were then screened in a mouse model (e) to identify the most potent vaccine (f). Structural model of T4-SARS-CoV-2 Nanovaccine showing an enlarged view of a single hexameric capsomer (g). The capsomer shows six subunits of major capsid protein gp23* (green), trimers of Soc (blue), and a Hoc fiber (yellow) at the center of capsomer. The expressible spike genes are inserted into phage genome, the 12 aa E external peptide (red) is displayed at the tip of Hoc fiber, S-trimers (cyan) are attached to Soc subunits, and nucleocapsid proteins (yellow) are packaged in genome core. See Results, Materials and Methods, and Supplementary Video for additional details.
For faster discovery of efficient vaccines against newly emerged pathogens such as SARS-CoV-2, it is crucial to develop a universal vaccine platform that can be incorporated with a variety of target antigens, such as DNA, proteins, peptides, and domains, either separately or in various combinations. Such a platform would be particularly helpful in designing and selecting the most appropriate vaccine without going through repetitive design cycles.
In the current study, the scientists developed a nanovaccine platform by bioengineering bacteriophage T4 using CRISPR technology. Such a platform can be used to rapidly produce vaccine candidates comprising multiple components of any emerging pathogen.
Study design
The scientists used bacteriophage T4 as a model platform to design vaccine candidates. The capsid of T4 is coated with two nonessential proteins namely Soc and Hoc. While Soc helps stabilize the capsid, Hoc helps T4 to attach to host cells. These two proteins can be used as adaptors to adhere antigenic determinants (epitope) to the capsid. Mechanistically, T4 capsid decorated with viral/bacterial epitopes acts as pathogen-associated molecular patterns, and thus can trigger the activation of pattern recognition receptors and induce strong immune responses.
To design the vaccine template, the scientists used SARS-CoV-2 as a model pathogen and inserted several viral components including the spike, envelop, and nucleocapsid as DNAs or proteins into T4 nanoparticle using CRISPR technology. To insert the viral components, they have removed the nonessential genomic regions of T4 phage. Importantly, they have used different compartments of T4 phage to insert different types of viral epitopes. Specifically, they have inserted full length spike gene, receptor binding domain gene, and nucleocapsid gene into the genome of T4 phage and co-packaged nucleocapsid molecules into the genome core. In addition, they engineered the T4 phage to display viral envelope peptide and spike-trimers.
By sequential engineering, they have created multiple recombinant T4 phages (vaccine candidates) comprising different combinations of pathogen epitopes.
To maintain the native-like structure of viral components, they have used SpyCatcher-SpyTag bridging approach, which is required for the appropriate folding and glycosylation of pathogen epitopes inside T4 phages.
Another important fact is that T4-based vaccines do not require adjuvant to induce desired immune responses. This helps reduce the cost of vaccine and complexity of manufacturing. Moreover, vaccines made without an adjuvant is generally safer than adjuvant-coupled vaccines because chemical adjuvants are often associated with vaccine-related reactogenicity. In this study, no adverse reactions have been noticed after vaccinating animals with T4-based vaccine candidates.
Immune responses
Upon testing these vaccine candidates in mouse and rabbit models, they researchers observed that the T4 phage displaying the viral spike trimers induces the highest levels of both antibody-mediated and T cell-mediated immune responses. The spike-specific antibodies are capable of blocking the interaction between the spike receptor binding domain and the host angiotensin converting enzyme 2 (ACE2) receptor, in addition to neutralizing the virus.
Taken together, this vaccine candidate is capable of providing complete protection against SARS-CoV-2 infection in mice. In addition to generating spike-specific antibodies, the vaccine induces a broad immune response against envelop and nucleocapsid proteins.
Study significance
The study has provided a T4 phage-based vaccine designing platform to cost effectively generate efficient vaccine candidates against any pathogen within a very short period. High levels of stability and safety profiles make bioengineered T4 a valuable platform to develop multipotent vaccine candidates comprising more than one distinct pathogen epitopes in a same formulation, which is important for generating robust and wide-spread immune responses.
Read the original article on News-Medical.net.