In modern science, nanopatterning is an essential technique for the fabrication of compact devices for electronic, optical, photonic, and biological applications. In this regard, ultraviolet nanoimprint lithography (UV-NIL) shows much promise owing to its low cost and scalability.
The technology is based on creating nanopatterns using UV light on a light-sensitive material called "resist" deposited on a substrate. After depositing the resist on the substrate, a mold nanopattern is pressed into the it. The resist fills this mold and is then cured using UV light, producing the desired nanopattern.
While UV-NIL is a well-explored technique, with simulations providing deep insights into the process, it is still limited to resolutions below 10 nm. This is because resolutions below 10 nm require an understanding of material features at atomic scales. Unfortunately, such feature cannot be explored with traditional simulations, which assume matter to be continuous.
While previous studies have looked at polymer-size effects on UV-NIL, behaviors of the short-chain resist molecules during the filling process remain unclear.
To address this issue, a research group led by Associate Professor Tadashi Ando from Tokyo University of Science (TUS), Japan, performed molecular dynamics (MD) simulations to elucidate the molecular features that govern the filling process at nanoscales. In their study published on July 25, 2022, in Nanomaterials, Dr. Ando and his colleague simulated the process of the filling of 2-nm and 3-nm mold trenches for four different resists, namely N-vinyl-2-pyrrolidone (NVP), 1,6-Hexanediol diacrylate (HDDA), Tri(propylene glycol) diacrylate (TPGDA), Trimethylolpropane triacrylate (TMPTA), and 2,2-Dimethoxy-2-phenylacetophenone (DMPA). Of these, HDDA, NVP, TPGDA, and TMPTA were photopolymers while DMPA was a polymerization initiator. Specifically, the team explored the effects of compositions and viscosities of these molecules on the UV-NIL filling process.
"The simulation results showed that HDDA, NVP/TPGDA/TMPTA, and TPGDA with viscosities lower than 10 mPa.s were able to fill the 2-nm and 3-nm trench widths, while the more viscous and bulkier TMPTA could not," highlights Dr. Ando. Specifically, molecules with viscosity higher than 92 mPa.s could not fill the trenches. Additionally, the researchers compared the two linear-shaped photopolymers, HDDA and TPGDA. The simulations revealed that TPGDA was relatively more flexible, making it more likely to undergo intramolecular crosslinking during UV-curing. Furthermore, these simulation results were in good agreement with empirical rules derived from experiments.
With these remarkable insights, the researchers are excited about the future prospects of UV-NIL. "The findings of our study could provide us useful information for guiding the future selection and design of optimized resists for fine nanopatterning at sub-10 nm resolution with UV-NIL," says Dr. Ando, excited.
We certainly hope his vision is realized soon!
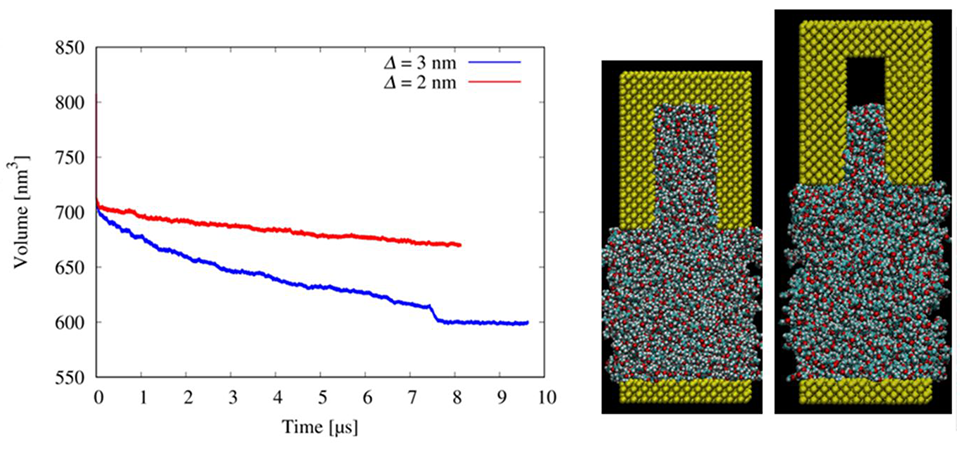
In a new study, researchers from TUS, use molecular dynamics simulations to understand the molecular features of resist materials that makes for better filling of nanometer-sized trenches in the nanopattern mold used in UV nanoimprint lithography.
Read the original article on Tokyo University of Science.