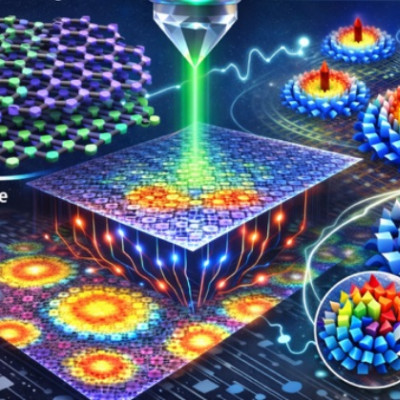
The new methods were applied to ligand-stabilized metal nanoparticles, which have been long studied at the Nanoscience Center at the University of Jyväskylä. Last year, the researchers published a method that is able to successfully predict binding sites of the stabilizing ligand molecules on the nanoparticle surface. Now, a new tool was created that can reliably predict potential energy based on the atomic structure of the particle, without the need to use numerically heavy electronic structure computations. The tool facilitates Monte Carlo simulations of the atom dynamics of the particles at elevated temperatures.
Potential energy of a system is a fundamental quantity in computational nanoscience, since it allows for quantitative evaluations of system’s stability, rates of chemical reactions and strengths of interatomic bonds. Ligand-stabilized metal nanoparticles have many types of interatomic bonds of varying chemical strength, and traditionally the energy evaluations have been done by using the so-called density functional theory (DFT) that often results in numerically heavy computations requiring the use of supercomputers.
This has precluded efficient simulations to understand nanoparticles’ functionalities, e.g., as catalysts, or interactions with biological objects such as proteins, viruses, or DNA. Machine learning methods, once trained to model the systems reliably, can speed up the simulations by several orders of magnitude.
The new method allowed simulations to be run on a laptop or desktop
In this work the researchers used the potential energies, predicted by the machine learning method, to simulate the atomic dynamics of thiol-stabilized gold nanoparticles. The results were in good agreement with the simulations performed by using the density functional theory. The new method allowed simulations to be run on a laptop or desktop in a time scale of a few hours while the reference DFT simulations took days in a supercomputer and used simultaneously hundreds or even thousands of computer cores. The speed-up will allow long-time simulations of the particles’ structural changes and particle-particle reactions at elevated temperatures.
The researchers used a distance-based machine learning method developed in the group of professor Tommi Kärkkäinen in Jyväskylä. It describes each momentary atomic configuration of a nanoparticle by calculating a so-called descriptor, and compares distances between descriptors in a multi-dimensional numerical space. By using correlations to a training set created by the reference DFT simulations, the potential energy can be predicted. This approach, used now for the first time in nanoparticle research, is simpler and more transparent than traditionally used neural networks.
“It is extremely motivating that we can reduce the computational load from running simulations in supercomputers to running them with similar quality in a laptop or a home PC”, says PhD student Antti Pihlajamäki who is the lead author of the study.
“It was a great surprise that our relatively simple machine learning methods work so well for complicated nanostructures”, states professor Tommi Kärkkäinen.
“In the next phase, our target is to generalize the method to work well for nanoparticles of many different sizes and chemical compositions. We will still need supercomputers to generate enough high-quality data to train the machine learning algorithm, but we hope that in the future we can move to use these new methods primarily to studies of nanoparticle functionality in complicated chemical environments”, summarizes Academy Professor Hannu Häkkinen, who coordinated the study.
The study was published in a Special Issue devoted to machine learning in The Journal of Physical Chemistry on May 15, 2020.
Read the original article on University of Jyväskylä.