Better tests needed
This development could cause many more false-negative diagnoses to arise, giving rise to a more devastating wave of community spread. The need for more up-to-date antibody arrays is obvious. A new preprint on the bioRxiv server reports the development of nanoantibodies (nAbs) – also called or nanobodiesTM or single-domain antibodies – that effectively bind the spike protein from the ancestral Wuhan variant as well as the recently detected UK and South African variants.
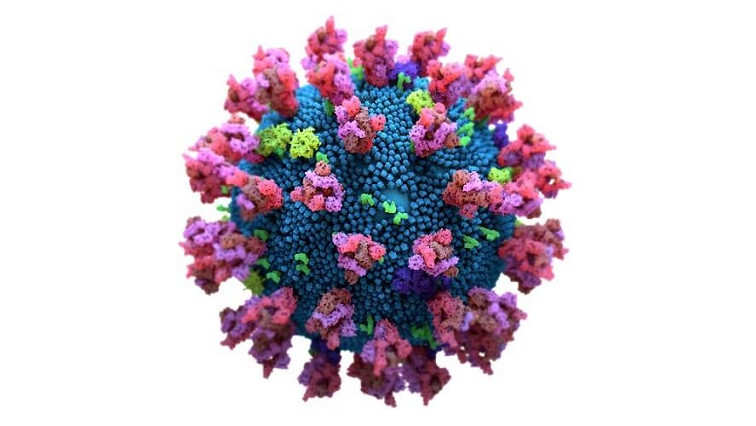
SARS-CoV-2 mediates host cell receptor attachment via its spike protein, which has two subunits, S1 and S2. The host cell receptor for the virus, the angiotensin-converting enzyme 2 (ACE2), is bound by the receptor binding domain (RBD) of the viral spike S1 subunit.
Mutational escape of detection
The UK variant, SARS-CoV-2 B.1.1.7 lineage, has approximately 20 mutations. Of these, the N501Y affects the RBD, while several are at the N-terminal domain of the S1 subunit of the spike protein. The South African variant, B.1.351 lineage, has a triple mutation in the RBD, namely, N501Y, K417N, and E484K.
These mutations may affect the epitope binding of the monoclonal antibody pairs that are currently used for antigen detection in rapid detection tests.
The benefits of nanobodies
Nanoantibodies overcome this obstacle by using a different mechanism. Since nAbs have a size ten times less than the classical immunoglobulin G (IgG) antibody, comprising a single domain, they creep into crevices on their cognate antigens by inserting the corresponding epitope-contacting antigens.
In contrast, conventional antibodies use a pair of arms comprising the heavy and light chains, to wrap around the epitope exposed on the surface.
The former mechanism of epitope binding allows greater flexibility in the face of individual amino acid substitutions, and has been exploited in the current study.
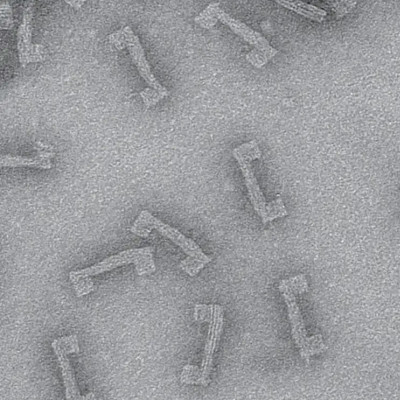
Llama nanobodies
The researchers first inoculated the SARS-CoV-2 spike S1 subunit and the receptor-binding domain (RBD) into a llama. They applied their already existing nanoantibody (nAb) development pipeline to obtain many groups of nAbs directed against the RBD. These showed strong binding for the RBD of the Wuhan-Hu-1 strain.
Each of these was then characterized. They found that two of them, nAb1 and nAb2, had strong binding affinity for the wildtype RBD.
Binding to all RBD variants
They created a preliminary version of a rapid antigen test, as a lateral flow assay (LFA), for testing against clinical samples predating the emergence of the newer variants. Using a fluorescent reporter protein, linked to a fusion protein containing two nAb1 domains (nAb1 dimer), they tested for binding affinity against the variant SARS-CoV-2.
They found that both nAb1 and nAb2 bound to RBDs from all three viral variants.
In contrast, the monoclonal antibody CR3022 binds to wildtype RBD only. Surprisingly, it failed to bind to the South African RBD variant, even though computational modeling predicted that the RBD would not be significantly affected by the variant's mutations.
nAb1 dimer shows higher binding
The nAb1 dimer has apparently higher binding affinity relative to the nAb1 monomer, perhaps because of cooperative binding, which may involve adjacent RBDs on the same immunobead. Since the natural form of the spike protein is trimeric, and each virion has about 24 trimeric spikes, cooperative binding is likely to boost the sensitivity of detection of the virus in clinical antigen detection kits.
In fact, earlier research by the same team has shown that linear multimers of nAb1 have a higher neutralizing capacity against SARS-Cov-2 infection of human iPSC-derived lung epithelial cells.
The researchers are continuing to test three or more nAb clusters for their ability to detect these variants and possibly others as well, currently in circulation or likely to emerge soon.
Highly sensitive detection
Already, Allele Biotech has produced a rapid antigen test for COVID-19 diagnosis for use at home and at the point of care. This device uses multiple nAbs to bind to hundreds of such binding pockets on the viral surface. This has added a high level of sensitivity, allowing current and future variants to be detected more confidently.
What are the implications?
The nanoantibodies for rapid antigen detection test bind the receptor binding domain (RBD) of the S1 protein from the original COVID-SARS-2 virus as well as those from the UK and South African variants. This finding validates our antibodies used in our assay for the detection of these major variant strains.”
Read the original article on Medical News.