DNA nanostructures have great potential for solving various diagnostic, therapeutic, and fabrication challenges due to their high biocompatibility and programmability. To function as effective diagnostic devices, for example, a DNA nanostructure might need to specifically respond to the presence of a target molecule by triggering an amplified read-out compatible with low-cost instruments accessible in point-of-care or clinical/laboratory settings.
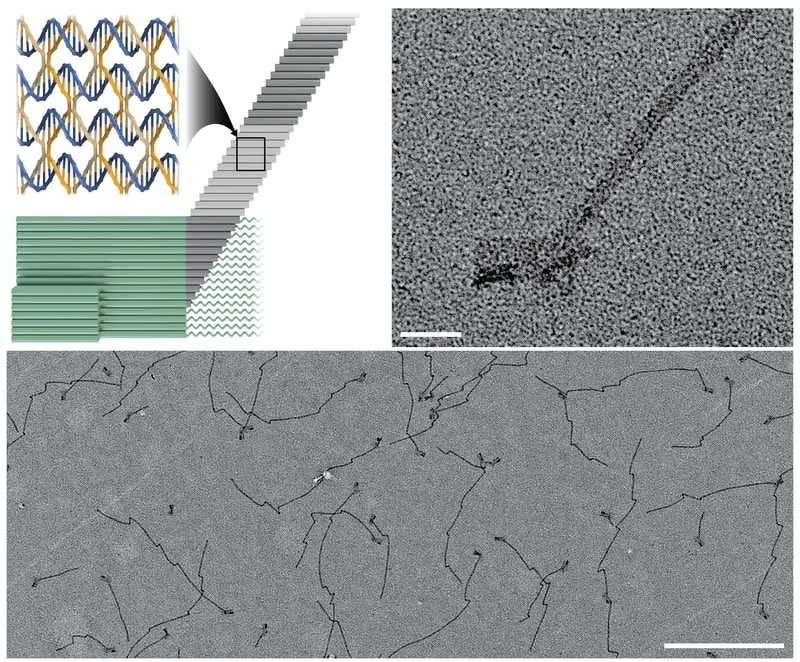
This diagram (top left) illustrates how in crisscross polymerization a “seed” structure (green) initiates an all-or-nothing assembly process at the nanoscale. The seeds exposes binding sites in the form of protruding single strands that can be detected by DNA “slats” (grey) weaving themselves into a continuously elongating nanoribbon that can become several micrometers long. The TEM images show a single tiny seed structure with a ribbon assembled on it (top right) at high magnification, and multiple elongated seed structures (bottom). Wyss Institute at Harvard University
Most DNA nanostructures are assembled using one of two main strategies that each have their strengths and limitations. “DNA origami” are formed from a long single-stranded scaffold strand that is stabilized in a two or three-dimensional configuration by numerous shorter staple strands. Their assembly is strictly dependent on the scaffold strand, leading to robust all-or-nothing folding. Although they can be formed with high purity in a broad range of conditions, their maximum size is limited. “DNA bricks” on the other hand can assemble much larger structures from a multitude of short modular strands. However, their assembly requires tightly controlled environmental conditions, can be spuriously initiated in the absence of a seed, and produces a significant proportion of incomplete structures that need to be purified away.
“The introduction of DNA origami has been the single most impactful advance in the DNA nanotechnology field over the last two decades. The crisscross polymerization approach that we developed in this study builds off this and other foundations to extend controlled DNA self-assembly to much larger length scales,” said Shih, who co-leads the Wyss’ Molecular Robotics Initiative, and also is Professor at Harvard Medical School and DFCI. “We envision that crisscross polymerization will be broadly enabling for all-or-nothing formation of two- and three-dimensional microstructures with addressable nanoscale features, algorithmic self-assembly, and zero-background signal amplification in diagnostic applications that require extreme sensitivity.”
We envision that crisscross polymerization will be broadly enabling for all-or-nothing formation of two- and three-dimensional microstructures with addressable nanoscale features, algorithmic self-assembly, and zero-background signal amplification in diagnostic applications that require extreme sensitivity.WILLIAM SHIH
Planting a seed
Having experienced the limitations of DNA origami and DNA brick nanostructures, the team started by asking if it was possible to combine the absolute seed-dependence of DNA origami assembly with the boundless size of DNA brick constructions in a third type of DNA nanostructure that grows rapidly and consistently to a large size.
“We argued that all-or-nothing assembly of micron-scale DNA structures could be achieved by designing a system that has a high free-energy barrier to spontaneous assembly. The barrier can only be bypassed with a seed that binds and arranges a set of ‘nucleating’ slats for joint capture of ‘growth’ slats. This initiates a chain reaction of growth-slat additions that results in long DNA ribbons,” said co-first author Dionis Minev, Ph.D., who is a Postdoctoral Fellow on Shih’s team. “This type of highly cooperative, strictly seed-dependent nucleation follows some of the same principles governing cytoskeletal actin or microtubule filament initiation and growth in cells.” The elongation of cytoskeletal filaments follows strict rules where each incoming monomer binds to several monomers that have previously been incorporated into the polymeric filament and in turn is needed for binding of the next one. “Crisscross polymerization takes this strategy to the next level by enabling non-nearest neighbors to be required for recruitment of incoming monomers. The resulting extreme level of coordination is the secret sauce,” said Minev.
This type of highly cooperative, strictly seed-dependent nucleation follows some of the same principles governing cytoskeletal actin or microtubule filament initiation and growth in cells. Crisscross polymerization takes this strategy to the next level by enabling non-nearest neighbors to be required for recruitment of incoming monomers. The resulting extreme level of coordination is the secret sauce.
From concept to actual structure(s)
Putting their concept into practice, the team designed and validated a system in which a tiny seed structure offers a high starting concentration of pre-formed binding sites in the form of protruding single DNA strands. These can be detected by DNA slats with six (or in an alternative crisscross system eight) available binding sites, each binding to one of six (or eight) neighboring protruding ssDNA strands in a crisscross pattern, and subsequent DNA slats are then continuously added to the elongating structure.
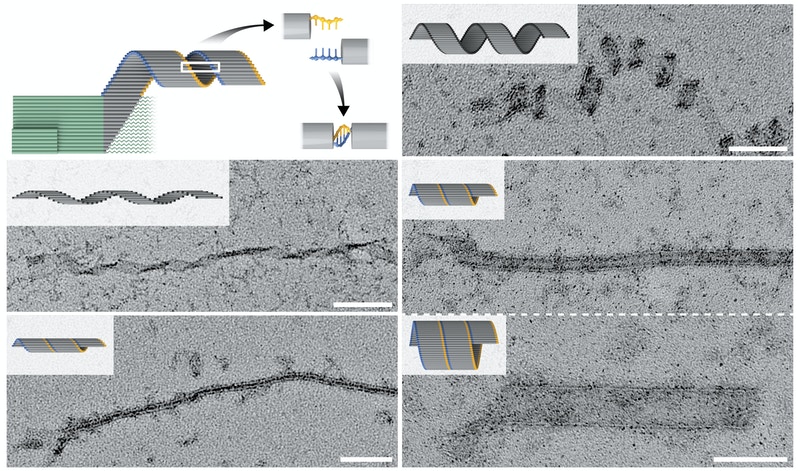
Strictly seed (green)-dependent crisscross polymerization enables the formation of diversely shaped tubes and coiled ribbons (grey), whereby elongating ribbons with different diameters are closed in different patterns by short complimentary single-stranded DNA overhangs (yellow and blue). This series of TEM images shows a variety of elongated nanoconstructs with the scale bar measuring 100 nanometers. Credit: Wyss Institute at Harvard University
“Our design is remarkable because we achieved fast growth of huge DNA structures, yet with nucleation control that is orders-of-magnitude greater than other approaches. It’s like having your cake and eating it too, because we readily created large-scale assemblies and did so only where and when we so desired,” said co-first author Chris Wintersinger, a Ph.D. student in Shih’s group who collaborated on the project with Minev. “The control we achieved with crisscross greatly exceeds that observed for existing DNA methods where nucleation can only be directed within a narrow window of conditions where growth is exceedingly slow.”
Using crisscross polymerization, Shih’s team generated DNA ribbons that self-assembled as a result of a single specific seeding event into structures that measured up to tens of micrometers in length, with a mass almost one hundred times larger than a typical DNA origami. Moreover, by leveraging the high programmability of slat conformations and interactions, the researchers created ribbons with distinct turns and twists, resulting in coiled and tube-like structures. In future studies, this could be leveraged to create functionalized structures that can benefit from spatially separated compartments. “An immediate application for our crisscross nanoconstruction method is as an amplification strategy in diagnostic assays following the formation of nanoseeds from specific and rare biomarkers,” said co-author Anastasia Ershova, who also is a Ph.D. student mentored by Shih.
Read the original article on Harvard University.