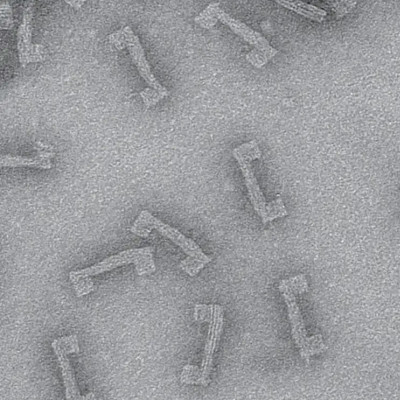
When harmful bacteria or viruses enter the body, immune cells spot telltale proteins known as antigens on the invaders’ surfaces and send out armies of antibodies to fend them off. If some of those antibodies have just the right shape, they can latch onto and block the antigens like the key to a padlock.
But our immune systems don’t always have the right antibodies to fight a particular invader. So over the past few decades scientists learned to work with animals such as camels and llamas, and to use synthetic design techniques in the lab, to generate antibodies that can be turned into medicines.
More than 85 antibody therapies have been approved by the FDA to date, including two granted emergency authorization for treating COVID-19.
Despite their success, current approaches have drawbacks. In an effort to leap these hurdles, researchers at Harvard Medical School and the University of California, Irvine, have developed a swifter, simpler, and cheaper adaptive technology to generate highly specialized antibodies.
They have already used the platform, dubbed AHEAD, to evolve antibodies against the virus that causes COVID-19. Other groups are now investigating those antibodies as the basis for diagnostic tests and therapies.
“We believe AHEAD will be a powerful tool for quickly discovering and optimizing antibodies, especially for addressing rapidly evolving pathogens,” said Andrew Kruse, professor of biological chemistry and molecular pharmacology in the Blavatnik Institute at HMS and co-senior investigator of the study with Chang Liu at UC Irvine.
Faster antibody discovery could accelerate drug development, diagnostic testing, and basic science experiments.
See inside a new technology that generates highly specialized antibodies for possible use in diagnostic tests and disease treatments.
As reported June 24 in Nature Chemical Biology, the method uses yeast to make hundreds of millions of different synthetic antibody fragments called nanobodies. Researchers can drop their antigen of interest—such as the spike protein that SARS-CoV-2 uses to enter and infect human cells—into a vial of the yeast and see which nanobodies latch on.
The team engineered the yeast so that the nanobodies evolve with each generation. That allows researchers to take the first-round winners, put them in a new vial, and conduct a second sort to get nanobodies that lock onto the antigen even more successfully. They can run additional rounds until they’re satisfied that they have one or more nanobodies that bind well, and bind only, to the disease-causing antigen, maximizing the chance of developing a therapy that is effective and has minimal side effects.
The whole process uses standard laboratory yeast culture techniques and takes just one and a half to three weeks. Researchers can hunt for nanobodies against many different antigens at the same time.
“We can evolve antibodies at previously inaccessible speed and scale,” said Kruse. “It’s a new way of doing combinatorial protein engineering.”
AHEAD is short for Autonomous Hypermutation yEast surfAce Display.
The work builds on an earlier platform led by Kruse and a colleague at the University of California, San Francisco. The new version differs in its autonomous evolution abilities, which mimic the way antibodies naturally evolve in llamas and camels.
“It’s exciting to bring this powerful immune process in animals to yeast cells,” said Conor McMahon, co-first author of the paper with Alon Wellner in the Liu lab. McMahon conducted the work while a postdoctoral fellow in the Kruse lab. He is now a Vertex Fellow at Vertex Pharmaceuticals.
Pandemic potential
While AHEAD has the potential to produce antibodies against threats such as cancers and proteins involved in autoimmune conditions, Kruse and colleagues are focused for the moment on using the technology to combat COVID-19.
“We wanted to get this project going as fast as we could,” said Kruse, “and we’re hoping we can now act even more quickly should something like this pandemic happen again.”
When the researchers introduced SARS-CoV-2 antigens into the yeast vials, they uncovered nanobodies that neutralized them at least as well as, and in some cases better than, existing antibodies generated from human patients, animals, and lab experiments.
The nanobodies had varying success in convincing the antigens to bind to them instead of to the ACE2 receptor, which SARS-CoV-2 uses to enter human cells.
Some colleagues who moved forward with the most promising nanobody candidates have begun to see similar results in animal models, while others are using the nanobodies to try to develop better tools to detect SARS-CoV-2 and related coronaviruses, according to Kruse and co-authors.
AHEAD could also help experts respond faster when new SARS-CoV-2 variants or entirely new pathogens arise.
“If SARS-CoV-2 evolves in a way that escapes current emergency-use antibody therapies, we should be able to evolve new ones in about two weeks to block the escape variants,” said Kruse.
Since “almost any biology lab” is equipped to use the simple equipment and techniques, AHEAD should empower many groups to work on finding solutions to future outbreaks “in a distributed response that meets the urgency of the problem,” Kruse added.
The labs of Debora Marks, assistant professor of systems biology at HMS, and Jonathan Abraham, assistant professor of microbiology at HMS, contributed to the work. The team published a related paper in Nature Communications detailing the novel computational techniques they developed to enable AHEAD.
Read the original article on Harvard University.