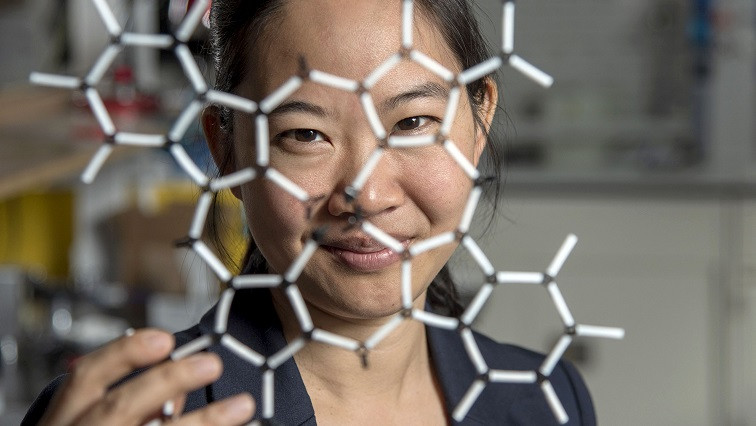
The Department of Materials Science and Engineering associate professor has teamed up with co-lead authors Blanka Janicek, a ’21 alumna and post-doc at Lawrence Berkeley National Laboratory (Berkeley Lab) in Berkeley, Calif., and Priti Kharel, a Department of Chemistry graduate student, to prove the methodology that allows researchers to visualize small molecular structures and accelerate current imaging techniques.
Additional co-authors include graduate student Sang hyun Bae and undergraduates Patrick Carmichael and Amanda Loutris. Their peer-reviewed research has recently been published in Nano Letters.
The team’s efforts expose the molecule’s atomic structure, allowing researchers to understand how it reacts, learn its chemical processes and see how to synthesize its chemical compounds.
“The structure of a molecule is so fundamental to its function,” Huang said. “What we’ve done in our work is make it possible to see that structure directly.”
The ability to see a small molecule’s structure is vital. Kharel shares just how vital by giving the example of a medication known as thalidomide.
Discovered in the ‘60s, thalidomide was prescribed to pregnant women to treat morning sickness and was later found to cause severe birth defects or, in some cases, even death.
What went wrong? The drug had mixed molecular structures, one responsible for treating morning sickness and the other unfortunately causing devastating, adverse effects to the fetus.
The need for proactive, not reactive science has urged Huang and her students to pursue this research effort that originally began with sheer curiosity.
“It’s so crucial to accurately determine structures of these molecules,” Kharel said.
Typically, molecular structures are determined with indirect techniques, a time-consuming and difficult approach that uses nuclear magnetic resonance or X-ray diffraction. Even worse, indirect methods can produce incorrect structures that give scientists the wrong understanding of a molecule’s makeup for decades. The ambiguity surrounding small molecules’ structures could be eliminated by using direct imaging methods.
In the last decade, Huang has seen significant advancements in cryogenic electron microscopy technology, where biologists freeze the large molecules to capture high-quality images of their structures.
“The question that I had was: What’s keeping them from doing that same thing for small molecules?” Huang said. “If we could do that, you might be able to solve the structure (and) figure out how to synthesize a natural compound that a plant or animal makes. This could turn out to be really important, like a great disease-fighter,” Huang said.
The challenge is that small molecules are often 100 or even 1,000 times smaller than large molecules, making their structures difficult to detect.
Determined, Huang’s students began using existing large molecule methodology as a starting point for developing imaging techniques to make the small molecules’ structures appear.
Unlike large molecules, the imaging signals from small molecules are easily overwhelmed by their surroundings. Instead of using ice, which typically serves as a layer of protection from the harsh environment of the electron microscope, the team devised another plan for keeping the small molecules’ structures intact.
How can you temper a molecule’s environment? By using graphene.
Graphene, a single layer of carbon atoms that form a tight, hexagon-shaped honeycomb lattice, dissipates damaging reactions during imaging.
Stabilizing the small molecule’s environment was only one issue the Illinois researchers had to manage. The team also had to limit its use of electrons, as low as one-millionth the number of elections normally used, to illuminate the molecules.
Low doses of electrons ensure that the molecules are still moving enough for the researchers to capture an image.
“The way I like to think about it is the molecule doesn’t like to be bombarded by higher-energy elections, but we need to do that to be able to see the structure, and graphene helps dissipate some of that charge away from the molecule so that we can actually get a nice image of it,” Janicek said.
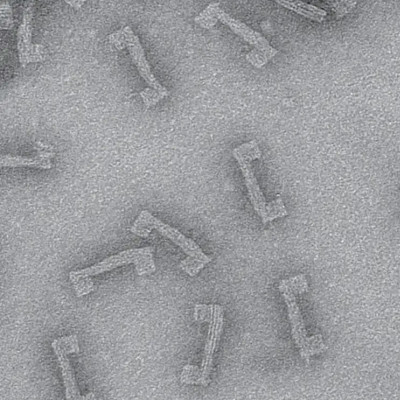
Unfortunately, once captured, the molecules were nearly invisible in the image.
“When they take a low-dose image, it initially looks like noise or TV static — almost like nothing is there,” Huang said.
The trick was to isolate the atomic structures from that noise by using a Fourier transform — a mathematical function that breaks down the small molecule’s image — to see its spatial frequency.
“We took images of hundreds of thousands of molecules and added them together to build a single, clear image,” Kharel said.
This averaging approach allowed the team to create crisp images of the molecules’ atoms without damaging the integrity of any individual molecule.
“Month after month, week after week, our resolution improved,” Huang said. “And then one day, my students came in and showed me the individual carbon atoms — that’s a major achievement. And of course, it comes after all this deep knowledge that they have gained to design an imaging experiment and how to unlock data from what looks like nothing.”
This collective discovery is paving the way for many more structural molecule imaging findings.
“There’s been this whole field of small molecules that have been left out in the cold, so to speak. We’re shining a light on how do we get there as a field? How do we make this thing that for us right now is so hard?” Huang said. “One day it won’t be — that’s the hope.”
The Illinois researchers’ efforts are the first big step in turning that dream into reality.
“One day, this will be the way we solve the structure of a small molecule,” Huang said. “People will simply throw the molecule in the electron microscope, take a picture and be done.”
That dream inspires Huang and her Illinois team to keep the course.
“That’s potentially life-changing, and we’ve made it exist,” Huang said. “We haven’t yet made it easy, but imaging techniques like this will change so much of science and technology.”
Read the original article on University of Illinois Urbana-Champaign.