Recent studies have shown that liquid-liquid phase separation – akin to how oil droplets form in water – leads to formation of diverse types of membraneless organelles, such as stress granules and nucleoli, in living cells. These organelles, also called biomolecular condensates, are liquid droplets performing specific cellular functions including gene regulation and stress response.
Now, a joint research team led by Professor Yongdae Shin and Do-Nyun Kim at Seoul National University announced that they harnessed the unique properties of the self-assembling DNA molecules to build synthetic condensates with programmable compositions and functionalities.
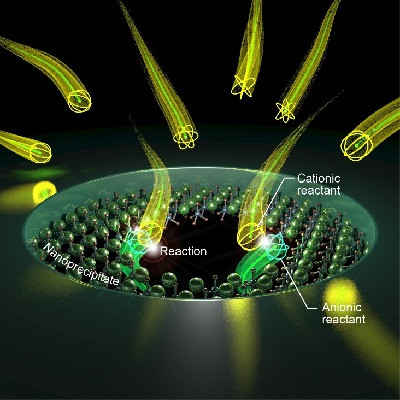
The researchers designed DNA scaffolds with motifs for self-association as well as specific recruitment of DNA targets. In a proper range of salt concentration and temperature, the engineered DNA scaffolds underwent liquid-liquid phase separation to form dense condensates, organized in a highly similar manner to those in living cells. The synthetic DNA condensates can recruit specific target DNA molecules, and the researchers demonstrated that the degree of recruitment can be precisely defined at the DNA sequence level.
They then endowed the synthetic condensates with functionalities by using DNA computation components as targets. DNA computing has been widely implemented for various bioengineering and medical applications, due to its intrinsic capacity of parallel computation. However, the slow speed of individual computation process has been a major drawback. With the synthetic DNA condensates, Shin and his team showed that DNA computation including logic gate operations were drastically sped up, by more than tenfold, when coupled to the condensates.
The architecture of DNA scaffolds also allowed selective recruitment of specific computing operations among many others running in parallel, which enabled a novel kinetics-based gating mechanism. The researchers expected that their system could be widely applied to diverse DNA circuits for disease diagnostics, biosensing, and other advanced molecular computations.
Read the original article on Newswise.