Electrochemical DNA biosensors hold significant promise for monitoring of various diseases. Overall, their detection applications are vast, from target DNA analytes such as bacterial genes and tumor sequences to clinically relevant concentrations of SARS CoV-2 biomarkers, for example. However, to achieve appropriate sensitivity and selectivity of such systems and enable their translation from the laboratory to a clinical environment is challenging, as these approaches often involve complex chemistries, electrochemical labeling, technically challenging materials, or multistep processing.
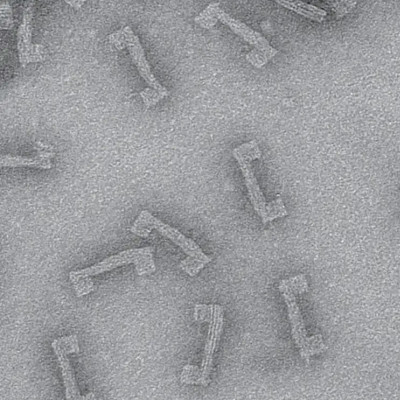
Now a team of researchers from Aalto University (Finland) and University of Strathclyde (Glasgow, UK) has found a way to significantly enhance the sensitivity of electrochemical DNA sensors by using modular DNA nanostructures as their novel components. The researchers combined the conventional DNA-based sensor techniques with programmable DNA origami structures to create a label-free sensor with significantly increased selectivity and sensitivity of detection.
‘In practice, our starting point is a rather simple and common DNA biosensor type – we have an electrode system immersed into the analyte solution, where the sensing electrode is coated with single-stranded DNA probe strands that are complementary to the (single-stranded) target DNA sequences. Once the target strand binds and hybridizes with the probe strand, the electric charges near the electrode move a bit, meaning that we can see a change in the electrochemical signal’, explains doctoral student Petteri Piskunen from Aalto University, one ofthe authors of the research.
‘Here DNA origami nanostructures come into play. We equipped our tile-like DNA origami with target-capturing strands that can efficiently and selectively bind to one end of the target sequence, while the other end of the target binds to the probe strands. Therefore, we are creating a sandwich-like complex, where the target strand is trapped between the electrode and the DNA origami. Then, instead of recording a small signal change upon the target binding, we will see an amplified effect due to the presence of the comparatively large DNA origami‘, continues Piskunen.
‘We demonstrated the feasibility of our system by detecting a gene fragment from bacteria with an antibiotic resistance. We could selectively trap this target from a rather complex solution that contained various types of single-stranded DNA, from short strands and junk fragments to long circular DNA. With our sensor we could reliably detect 100–1000 times lower target concentrations than with the conventional techniques´, says Visiting Scientist Veikko Linko (currently Associate Professor at University of Tartu, Estonia).
‘It is encouraging to think that by combining versatile DNA origami with for example printable and disposable electrodes, we could create label-free sensing platforms with such high sensitivity and specificity. This positions our technology with a route to mass manufacturability and broad applicability as point-of-care devices. At the moment, collaborative work with the University of Strathclyde is ongoing for generalizing the sensor setup for use with different kinds of biomarkers’, Linko concludes.
Read the original article on Aalto University.