Isolating low levels of proteins
The analysis of proteins in biological systems by mass spectrometry has become well established, whether for human, animal, botanical, or bacterial systems, but there remain aspects that have need of improvement. One of these areas involves samples that contain especially low amounts of protein that need special workflows to enrich the proteins and improve proteome coverage. This issue has been examined by a research team from Shanghai Jiao Tong University, China, as described by Xianting Ding.
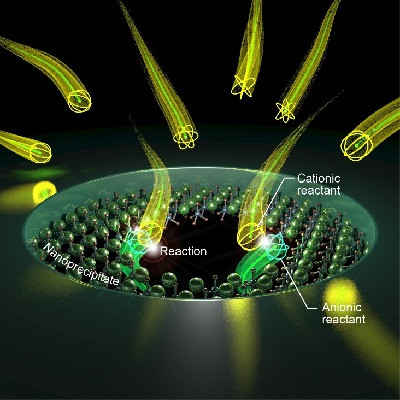
They developed a novel functional material to trap trace proteins that reduced sample losses and avoided the co-extraction of other biocompounds such as nucleic acids, salts, and lipids and was also amenable to automation. The trapping compounds, known as molecular glues, were cucurbit[7]uril and sulfonated calix[8]arene. They were covalently attached in turn to superparamgnetic iron oxide (Fe3O4) nanoparticles that had been surface-functionalized with a titanium dioxide layer.
In tests and docking simulations with model proteins, a range of noncovalent interactions were found to be involved in trapping, depending on the particular protein. Hydrophobic interactions, electrostatic attraction, and Van der Waals interactions were the main forces.
Molecular glues in streamlined wokflow
In a test of their practical application, the functionalized nanoparticles were incorporated into a scheme, named the Streamlined Workflow based on Anchor-nanoparticles for Proteomics (SWAP), for extracting proteins from solution. It was demonstrated with whole-cell lysates of HEK293 and HeLa cells and the whole process, from sample loading to trypsin digestion and elution, took place in a single tube.
The nanoparticles were added to the lysates and vortexed for five minutes to encourage binding before a magnet was brought to the tube to retain the nanoparticles as the supernatant was poured off. After rinsing with ammonium carbonate solution, formic acid solution was added to the tube to release the proteins which were digested with trypsin overnight. The resulting peptide solution was collected for analysis by mass spectrometry.
For samples containing 106 cells, more than 6000 proteins were identified with 79.6% coverage. As the number of initial cells was reduced, so did the number of proteins but lots with as few as 100 cells still yielded more than 2400 proteins.
In direct comparison with two established techniques – filter-aided sample preparation (FASP) and surfactant-assisted one-pot processing (SOP) – SWAP outperformed them both in terms of sensitivity, reproducibility, qualitative, and quantitative performance. This was achieved by minimizing sample losses throughout the sample handling process.
Eyeing proteins in aqueous humor
The SWAP system was applied to real human samples of aqueous humor which contains low protein levels. For just 5 µL of aqueous humor, the SWAP protocol outperformed FASP, yielding an average of 1204 proteins compared with 1077, signaling fewer protein losses during processing. For three parallel experiments, the number of overlapping proteins identified in the three sets was also greater for SWAP, being 93.3% versus 82.0%.
In a further demonstration of SWAP, it was applied to the proteomics of aqueous humor of patients with wet age-related macular degeneration (wAMD) or cataracts. A total of 1171 proteins were identified from both sets, with 31 and 100 proteins expressed only on wAMD or cataract patients, respectively. Certain proteins involved in eye protection were reduced in abundance for wAMD compared with cataracts.
The experiments were repeated following treatment with the drug aflibercept, which is designed to challenge the overexpression of the protein vascular endothelial growth factor (VEGF). Proteomics analysis using the SWAP system confirmed a decrease in VEGF levels, as well as those of other proteins involved in inflammation.
The functionalized nanoparticles containing molecular glues, combined with the one-pot SWAP system minimized sample losses by reducing protein adsorption and restricting rinsing and filtering steps and was beneficial to the analysis of biosamples containing low levels of protein.
Read the original article on Wiley Analytical Science.