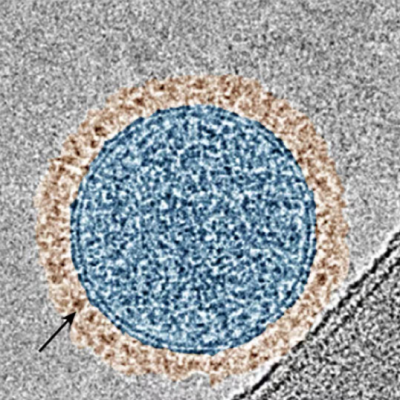
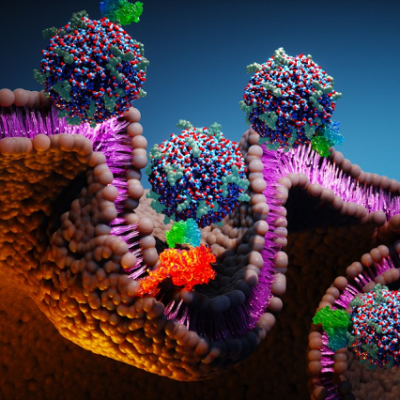
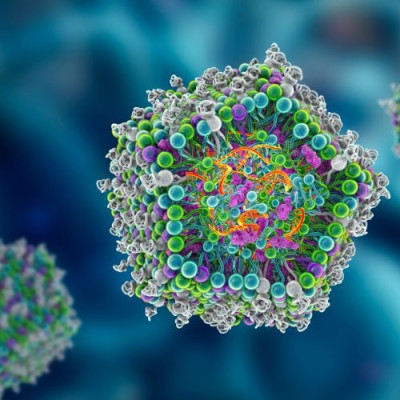
Scientists from NPL, working in collaboration with AI experts from IBM and the Science and Technology facilities Council (STFC) Hartree Centre, are applying the principles of precision engineering of biological systems to enable the design of artificial virus-like particles capable of encapsulating genes. Such particles, known as virions, can be used to deliver desired genetic material into cells with broad ranging applications including gene therapy and engineering biology. Delivering drugs specifically to the required site of action poses substantial challenges, and is key to drug development where scientists seek to enable a desired therapeutic effect without causing negative side-effects - this work has the potential to contribute solutions to this challenge.
This new study, published in ACS Nano, addresses specific challenges in virion design using approaches that are not observed in virus biology. The approach uses naturally occurring amino acids - the building blocks of proteins. Specifically, the virions are formed from (L-amino acids) assembling alternately with their mirror reflections, known as D-amino acids which assemble to create virus-like shells which can be designed to enclose nucleic acids of different sizes. The work will have a wide range of applications but is currently being exploited most notably in applications such as personalised medicine and gene therapy. Alternative synthetic virion shells have also been shown to have antimicrobial activity providing potential for development of a new class of antibiotic alternatives.
The study provides an excellent example of predictive biological design making use of AI models able to support prediction of both the number of molecules necessary to assemble into each virion, and the amount of nucleic acid captured in each virion.
Read the original article on National Physical Laboratory (NPL).