SARS-CoV-2, the new coronavirus behind the COVID-19 pandemic, infects cells by using its spike proteins to attach to proteins present on the surfaces of certain human cells, also known as ACE2 receptors. Upon binding to ACE2, the virus fuses with the host cell membrane and gains entry via this attachment.
Finding inhibitors that block key regions of the spike protein and keep the virus from infecting cells is the focus of many scientists from around world.
At the University of Illinois at Chicago (UIC), researchers recently published a study in the journal ACS Nano that details findings from computer simulations seeking to identify these inhibitors, which eventually could assist chemists to develop new medicines to combat the coronavirus.
Using a recently published X-ray crystal structure of the receptor-binding domain of SARS-CoV-2 when it is bound to ACE2, the researchers were able to identify 15 amino acids from ACE2 that interact directly with the viral spike protein.
Yanxiao Han, a UIC Ph.D. student in chemistry and first author on the paper, and Petr Král, UIC professor of chemistry, physics, biopharmaceutical sciences and chemical engineering, used computer modeling to design and assess four peptides that mimic the virus-binding domain of the human protein that allows SARS-CoV-2 to enter cells.
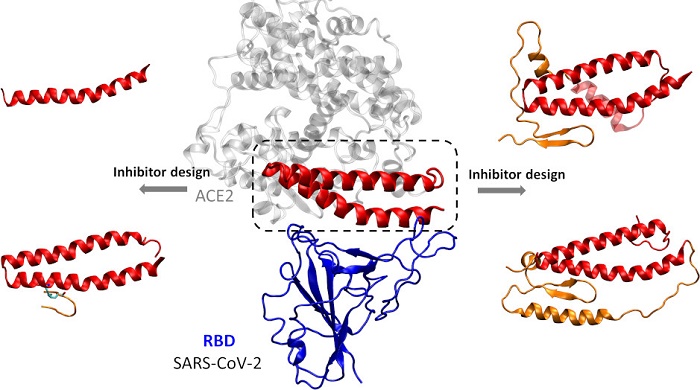
Computational design of ACE2-based peptide inhibitors of SARS-CoV-2. Photo: Yanxiao Han and Petr Král
Their molecular dynamics simulations revealed a double alpha helix extracted from ACE2 as a highly stable peptide and potential defender from the virus, capable of blocking the spike proteins.
“A single alpha helix was not particularly stable,” said Král, senior author of the study. “So, the discovery that a pair of linked self-supporting helices can provide strong and stable binding was very exciting.”
Han and Král hope that simple peptides might do the job of healing the patients in later stages by slowing down the activity of the virus.
“Such peptides might have limited secondary impact on the organisms,” Král said. “These are not vaccines but possible therapeutics for inhalation.”
The peptides still need to be tested in the lab and in patients, but being able to narrow down drug candidates on the computer could help advance this process, according to the researchers. Han’s work related to the study is supported through the UIC Graduate College Dean’s Scholar Fellowship.
Read the original article on University of Illinois at Chicago.