

| Date | 27th, Dec 2018 |
|---|
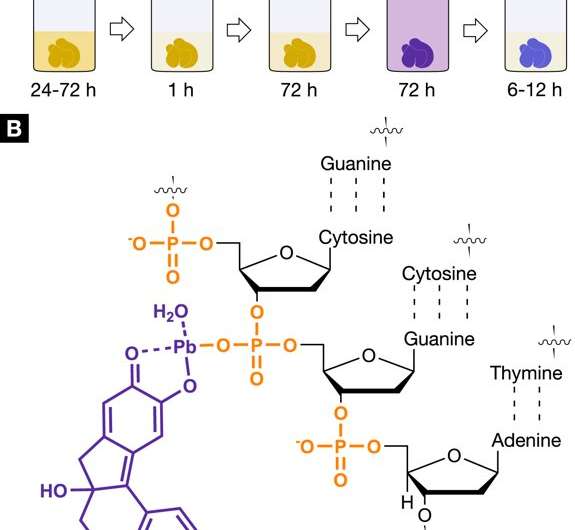 The X-ray suitable, nucleus-specific staining agent protocol for 3D virtual histology. Staining protocol and interaction of the hematein-based X-ray stain with soft tissue. (A) The developed hematein-based staining procedure shows the individual steps involved including incubation and staining times. Staining step 1 was conducted using lead(II) acetate trihydrate as the heavy metal source. The lead(II) acetate trihydrate was dissolved in distilled water and is referred to as working solution (A) (WS (A)). The staining step 2 involved a hematein solution in absolute ethanol (WS (B), 10% (w/v); c = 333 mM), which was derived from hematoxylin and was added to WS (A). (B) The positively charged hematein lead(II) complex (purple), which is built in situ in the soft-tissue sample, is interacting with the negatively charged phosphate backbone of the DNA (orange) present in the nucleus of the cell. The selective interaction of the hematein lead(II) complex with the DNA is achieved by acidification of the soft tissue during fixation or before staining and allows for a higher accumulation of the hematein lead(II) complex within the cell nucleus. Credit: Scientific Reports, doi: https://doi.org/10.1038/s41598-018-36067-y
The X-ray suitable, nucleus-specific staining agent protocol for 3D virtual histology. Staining protocol and interaction of the hematein-based X-ray stain with soft tissue. (A) The developed hematein-based staining procedure shows the individual steps involved including incubation and staining times. Staining step 1 was conducted using lead(II) acetate trihydrate as the heavy metal source. The lead(II) acetate trihydrate was dissolved in distilled water and is referred to as working solution (A) (WS (A)). The staining step 2 involved a hematein solution in absolute ethanol (WS (B), 10% (w/v); c = 333 mM), which was derived from hematoxylin and was added to WS (A). (B) The positively charged hematein lead(II) complex (purple), which is built in situ in the soft-tissue sample, is interacting with the negatively charged phosphate backbone of the DNA (orange) present in the nucleus of the cell. The selective interaction of the hematein lead(II) complex with the DNA is achieved by acidification of the soft tissue during fixation or before staining and allows for a higher accumulation of the hematein lead(II) complex within the cell nucleus. Credit: Scientific Reports, doi: https://doi.org/10.1038/s41598-018-36067-y
Histology is used to identify structural details of tissue at the microscale in the pathology lab, but analyses remain two-dimensional (2D) as they are limited to the same plane. Nondestructive 3D technologies including X-ray micro and nano-computed tomography (nanoCT) have proven validity to understand anatomical structures, since they allow arbitrary viewing angles and 3D structural detail. However, low attenuation of soft tissue has hampered their application in the field of 3D virtual histology. In a recent study, now published on Scientific Reports, Mark Müller and colleagues at the Department of Physics and Bioengineering have developed a hematein-based X-ray staining method to specifically target cell nuclei, followed by demonstrations on a whole liver lobule of a mouse.
The novel staining protocol combined the recently developed, high resolution nanoCT system for 3D visualization of the tissue architecture at the nanometer scale. The results revealed the real 3D morphology alongside spatial distribution of cell nuclei. The technique was also compatible with conventional histology, as microscopic slides with soft tissue sample could be stained with the same protocol alongside additional counter-staining. The method demonstrated the possibility for future applications in histopathology accompanied with X-ray CT devices in the lab.
Histology is the existing gold standard for an accurate microanatomical diagnosis in the pathology lab, however techniques and results are limited to 2D. For instance, a 3D biopsy is usually examined using very thin microscopic slides (containing 2-10 µm thick slices) via conventional and modern immunohistochemical and histological staining techniques. Micro- and nano-CT are powerful tools that can provide an accurate reconstruction of tissue in 3D. Developments on the technology have allowed comparatively high resolution to existing 2D conventional histology, using devices ranging from large particle accelerators to laboratory-based X-ray devices.
Alongside the technical requirements, X-ray suitable staining agents (contrast agents) such as phototungstic acid (PTA), iodine potassium iodide (IKI), iodine in ethanol (12E), or iodine in methanol (12M) are also important. The available staining agents are however, currently limited in scope and efficacy. When developing next-generation medical diagnostics in histopathology, scientists aim to optimize the techniques and understand tissue architecture from the cellular level to the tissue scale.
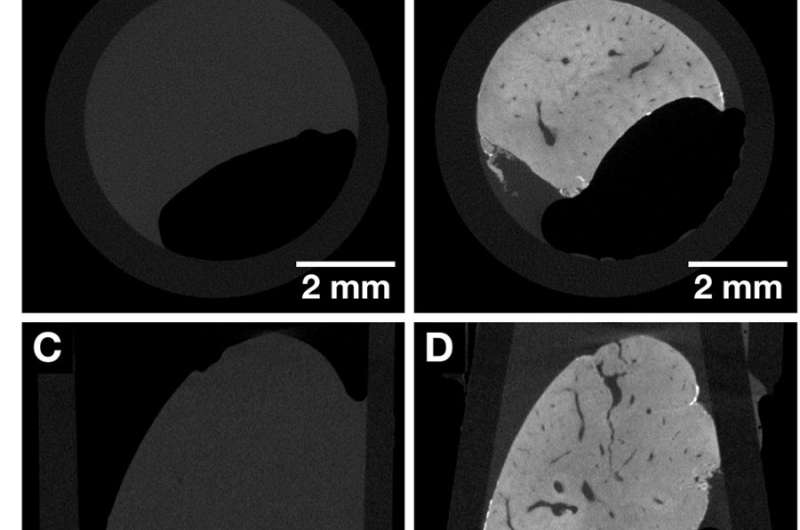 CT slices of the same whole mouse liver lobule before and after staining highlighting the contrast enhancement obtained after application of the hematein-based X-ray stain. Both data sets were acquired with the Xradia Versa 500 microCT using identical acquisition parameters. The voxel size in both data sets is 13.5 µm. (A, C and E) Overview images of the unstained mouse liver lobule representing the views along the Cartesian axes. (B, D and F) Overview images of the same mouse liver lobule sample in (A, C and E) after staining representing the views along the Cartesian axes. Anatomical structures such as the vasculature are visualized. Credit: Scientific Reports, doi: https://doi.org/10.1038/s41598-018-36067-y.
CT slices of the same whole mouse liver lobule before and after staining highlighting the contrast enhancement obtained after application of the hematein-based X-ray stain. Both data sets were acquired with the Xradia Versa 500 microCT using identical acquisition parameters. The voxel size in both data sets is 13.5 µm. (A, C and E) Overview images of the unstained mouse liver lobule representing the views along the Cartesian axes. (B, D and F) Overview images of the same mouse liver lobule sample in (A, C and E) after staining representing the views along the Cartesian axes. Anatomical structures such as the vasculature are visualized. Credit: Scientific Reports, doi: https://doi.org/10.1038/s41598-018-36067-y.
During the first diagnosis in clinical pathology, the cell nuclei and cytoplasm are of significance. Almost every histological sample is therefore often stained via the standard hematoxylin and eosin (H&E) protocol to identify the nuclei and cytoplasm. But standards for the hematoxylin stain are not fixed and many variants to the protocol exist due to diverse tissue types and/or pre-treatment parameters involved. As a result, Müller et al. introduced a hematein-based staining protocol, specifically developed for CT, to allow for direct 3D visualization of cell nuclei in soft-tissue samples. The powerful potential of microCT or nanoCT combined with X-ray suitable stains will allow future insights into tissue organization to understand disease including osteoarthritis and cancer, at the cellular nanoarchitecture.
When developing the new X-ray suitable, hematein-based stain, the commonly used Mayer's hematoxylin and Wiegert's iron hematoxylin were included in its constitution. The staining experiments were conducted on the tissue of mouse liver lobule, used for X-ray CT-imaging thereafter. The optimized staining protocol contained five steps starting with controlled acidification of soft-tissue samples during fixation. The soft-tissue was prepared on the molecular level to stain with the hematein lead (II) complex.
In its mechanism of action, the scientists showed strengthened ionic interaction between the positively charged hematein lead (II) complex and the negatively charged phosphate backbone of the deoxyribonucleic acid (DNA). The chemical reaction ensured higher accumulation of the staining agent within the cell nuclei by forming a hematein lead (II) DNA complex, as an X-ray suitable agent.
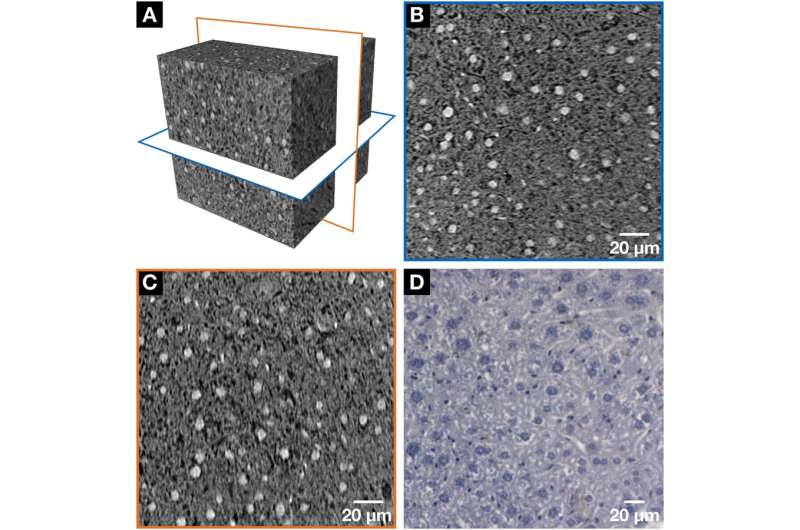 NanoCT data (A–C) in comparison with the histological microscopic slide (D) derived from the same mouse liver lobule after application of the hematein-based X-ray staining protocol. Clear visualization of the larger hepatocyte cell nuclei and the smaller cell nuclei such as Kupffer cells and SECs in white (A–C) or dark purple (D) and the BC network displayed in black (A-C) or white (D) was achieved, respectively. (A) The volume of interest (VOI) highlighting the two nanoCT slices shown in (B, blue frame) and (C, orange frame). (B, C) Representative individual nanoCT slices as indicated in the VOI from (A). (B) and (C) are positioned orthogonal to each other. The orientation of the BC network, which is formed by the hepatocytes is seen, i.e. more horizontal arrangement is seen in (B) and a more vertical alignment in (C). The nanoCT slice thickness is 580 nm. (D) Representative histological microscopic slide with a thickness of 3 µm obtained from the same mouse liver lobule sample after applying the hematein-based stain and embedding it in a paraffin block. Credit: Scientific Reports, doi: https://doi.org/10.1038/s41598-018-36067-y.
NanoCT data (A–C) in comparison with the histological microscopic slide (D) derived from the same mouse liver lobule after application of the hematein-based X-ray staining protocol. Clear visualization of the larger hepatocyte cell nuclei and the smaller cell nuclei such as Kupffer cells and SECs in white (A–C) or dark purple (D) and the BC network displayed in black (A-C) or white (D) was achieved, respectively. (A) The volume of interest (VOI) highlighting the two nanoCT slices shown in (B, blue frame) and (C, orange frame). (B, C) Representative individual nanoCT slices as indicated in the VOI from (A). (B) and (C) are positioned orthogonal to each other. The orientation of the BC network, which is formed by the hepatocytes is seen, i.e. more horizontal arrangement is seen in (B) and a more vertical alignment in (C). The nanoCT slice thickness is 580 nm. (D) Representative histological microscopic slide with a thickness of 3 µm obtained from the same mouse liver lobule sample after applying the hematein-based stain and embedding it in a paraffin block. Credit: Scientific Reports, doi: https://doi.org/10.1038/s41598-018-36067-y.
To compare the staining efficacy, the mouse liver lobule tissue was imaged with microCT before staining, followed by imaging after hematein-protocol based staining. The staining process occurred in two steps and the desired contrast enhancement was achieved as expected after staining. The results were observed using the microCT overview to show distinct anatomical structures, including the vasculature. The staining was homogenous within the entire mouse liver lobule, unlike with previous examples in large liver tissue samples. The 3D imaging process allowed accessibility to a series of CT slices in arbitrary planes. Furthermore, unlike conventional 2D histology (with paraffin-embedded soft-tissue), the soft-tissue could be viewed from different angles.
The tissue was next investigated at the sub-cellular level with smaller pieces of the same liver lobule, which were dissected and analyzed using nanoCT. The visualized results showed regions of the volume of interest (VOI), cell nuclei of the hepatocytes and nuclei of other cell types (Kupffer cells and sinusoidal endothelial cells). Black whole structures represented the bile canalicular (BC) network while the darker gray values indicated the cytoplasm of the liver tissue sample. The orientation of the BC network was also observed. The tissue was then investigated with conventional histology, using very thin microscopic slides with no other staining aside from the hematein-based stain applied. The conventional technique similarly confirmed the morphology of hepatocytes and of other cell types (nuclei stained in dark purple), while the BC network was stained in white.
 3D nuclei and analysis of different cell nuclei present in the mouse liver volume of interest (VOI). Credit: Scientific Reports, doi: https://doi.org/10.1038/s41598-018-36067-y.
3D nuclei and analysis of different cell nuclei present in the mouse liver volume of interest (VOI). Credit: Scientific Reports, doi: https://doi.org/10.1038/s41598-018-36067-y.
The scientists comparatively confirmed 3D data reconstruction accuracy in the study with previous investigations. When the hematein-based procedure was re-applied in conventional 2D histology investigations, the scientists also applied a standard counter-stain, eosin Y specific, to the cell cytoplasm on to the tissue. In the results, the cell nuclei remained purple while the cytoplasm took up the pink stain. In this way, the scientists authenticated the nuclei-specific hematein-based staining capacity with standard histology as well.
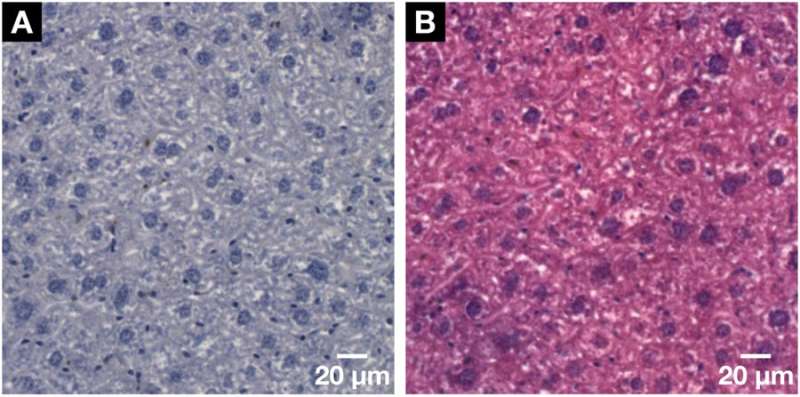 Demonstration of the histological compatibility of the for X-ray microCT and nanoCT developed hematein-based staining method with conventional 2D histology. (A) Representative histological microscopic slide with a thickness of 3 µm obtained from the same mouse liver lobule sample after the applied hematein-based staining and embedding in a paraffin block. Clear visualization of the larger hepatocyte cell nuclei and the smaller cell nuclei such as Kupffer cells and SECs in dark purple and the BC network displayed in white. (B) The compatibility with the standard counter stain of eosin Y was shown on a subsequent microscopic slide seen in (A). The cell nuclei are shown in purple next to the cytoplasm in pink resulting in a typical H&E stained microscopic slide of a soft-tissue sample. Credit: Scientific Reports, doi: https://doi.org/10.1038/s41598-018-36067-y.
Demonstration of the histological compatibility of the for X-ray microCT and nanoCT developed hematein-based staining method with conventional 2D histology. (A) Representative histological microscopic slide with a thickness of 3 µm obtained from the same mouse liver lobule sample after the applied hematein-based staining and embedding in a paraffin block. Clear visualization of the larger hepatocyte cell nuclei and the smaller cell nuclei such as Kupffer cells and SECs in dark purple and the BC network displayed in white. (B) The compatibility with the standard counter stain of eosin Y was shown on a subsequent microscopic slide seen in (A). The cell nuclei are shown in purple next to the cytoplasm in pink resulting in a typical H&E stained microscopic slide of a soft-tissue sample. Credit: Scientific Reports, doi: https://doi.org/10.1038/s41598-018-36067-y.
The hematein-based staining protocol aided high resolution CT visualization for cell nuclei in soft tissue at the sub-micron range, hitherto not possible with other staining methods that were combined with microCT technology. Future histopathological studies may be able to eliminate time-consuming preparation procedures and tissue sample loss (as seen with standard histology) to obtain individual tissue slices via 3D investigation of an entire VOI, as demonstrated in the present study. The ability to screen a larger sample for abnormal cell nuclei could assist pathologists to identify regions of inflammation to assess disease etiology and progression.
The staining protocol is simple and reproducible, suited for whole-organ CT staining, coupled with 3D visualization and in-depth, non-destructive analysis of soft tissue samples. The staining agents listed in the protocol are easily accessible, while the CT combined X-ray suitable protocol allows greater sophistication for soft-tissue sample analyses. Steps in the staining protocol will require further optimization with different tissue types and in diverse applications, including 3D histology, developmental and structural biology studies in the lab.
More information: 1. Mark Müller et al. Nucleus-specific X-ray stain for 3D virtual histology, Scientific Reports (2018). DOI: 10.1038/s41598-018-36067-y
2. Juliana Martins de S. e Silva et al. Three-dimensional non-destructive soft-tissue visualization with X-ray staining micro-tomography, Scientific Reports (2015). DOI: 10.1038/srep14088
3. X-ray microtomographic imaging of intact vertebrate embryos www.ncbi.nlm.nih.gov/pubmed/22135670 , Metscher B.D. et al. December 2011, Cold Spring Harbor Protocol.
4. Three-dimensional virtual histology enabled through cytoplasm-specific X-ray stain for microscopic and nanoscopic computed tomography www.ncbi.nlm.nih.gov/pubmed/29463748?dopt=Abstract, Busse M. et al. March 2018, Proceedings of the National Academy of Sciences
© 2018 Science X Network
Citation: Nucleus-specific X-ray stain for 3-D virtual histology (2018, December 27) retrieved 22 August 2022 from https://phys.org/news/2018-12-nucleus-specific-x-ray-d-virtual-histology.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without the written permission. The content is provided for information purposes only.
