

| Date | 6th, Jan 2022 |
|---|
Protein assembly is essential for the formation of ordered biological structures, but imagine engineering one! This is exactly what researchers at Tokyo Tech have now accomplished with protein needles. By regulating the tip-to-tip interactions of these needles, they allowed for their self-assembly into lattice structures, ordered monomeric states, and fiber assemblies, paving the way for the controlled construction of more of such protein architectures.
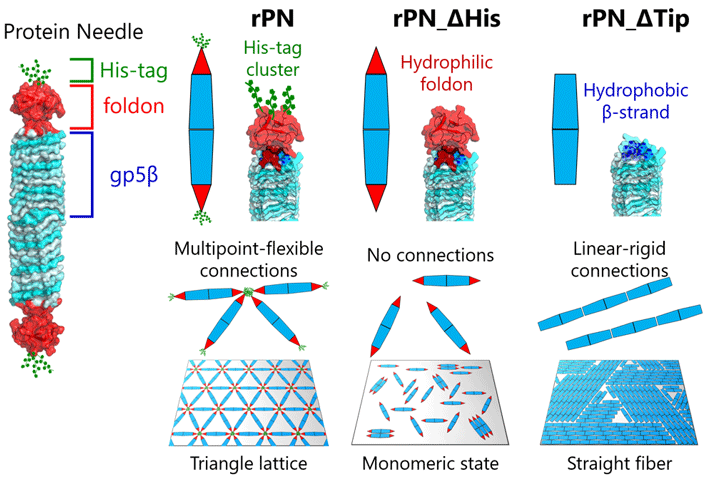
Figure 1. Engineered protein needles and their assembly on a mica surface
Scientists have long been attempting to decode the complex sub-structures of proteins. Now, researchers from Tokyo Tech have finally shed light on this front with the investigation of engineered protein self-assembly using protein needles.
Proteins are the basic building blocks of our bodies. However, their molecular and macroscopic structures are complex and varied, with multiple folding patterns and substructures. Scientists have been trying to decode these structures for some time, and much progress has been made thanks to fluorescence microscopy (FM), atomic force microscopy (AFM), and high-speed AFM (HS-AFM). However, they have not been able to directly observe the dynamic motions of proteins during assembly. This is mainly due to the intricate structure of proteins, which are too small to be measured with existing techniques.
A collaborating team of researchers from Tokyo Institute of Technology (Tokyo Tech), Kyushu University, Nagoya University, and National Institutes of Natural Sciences have now developed a specialized anisotropic protein needle (PN) to help determine the assembly of similarly anisotropic proteins, giving us clues about their microstructure and assembly.
Prof. Takafumi Ueno of Tokyo Tech, who led the study, explains the premise of their work, "Our PN is a needle-shaped protein composed of the rigid body (β-helix), the terminal cap (foldon), and a binding motif (hexa-histidine tag, His-tag). By modifying these PNs by deleting the His-tag motif and foldon cap, we can produce three different types of PNs. This enabled us to regulate and observe different assembly patterns and how they change, giving us clues into the mechanics of different protein-protein interactions that we find in nature." The results of this study were published in the journal Small.
In solution, the PNs spontaneously form a highly stable structure with a length of about 20 nm and a width of about 3.5 nm, small enough to track the rotational motion of individual molecules yet mechanically strong.
On surfaces, the team observed different kinds of ordered structures as the PNs self-assembled. These structures ranged from triangular lattices and monomeric states with nematic order (one-dimensional orientation) to fiber assemblies (Figure 1).
This, in turn, allowed the team to investigate the dynamic processes involved in protein assembly through a combination of HS-AFM and simulations (Figure 2). The results revealed that the formation of the triangular lattice structure was guided by the dynamic motions of PN, which contribute to forming ordered lattices (Figure 3).
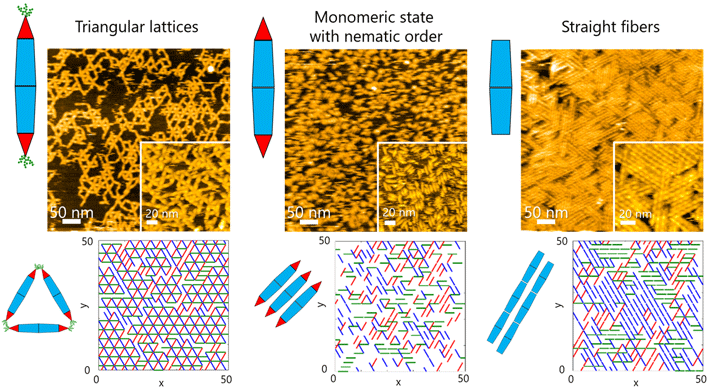
Figure 2. High-speed atomic force microscopy observations and Monte Carlo simulations of two-dimensional self-assembly
Basic protein chains often undergo dynamic self-assembly to form complex supramolecular structures. Scientists at Tokyo Tech have now managed to explore the assembly dynamics using engineered protein needles.
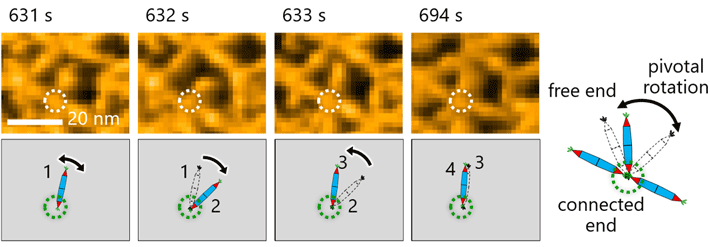
Figure 3. Molecular motion of the protein needles (rPN) showed pivotal rotations around the His-tag interaction
Observing the molecular motion of the PNs was crucial for making observations in this study. Here, researchers noted pivotal rotation around the His-tag interaction between protein needles.
These findings have excited the researchers, who are contemplating its potential ramifications. "These molecules play such a crucial role in biological systems that understanding their structure would further the field significantly. For instance, we could use this to lay the groundwork for constructing supramolecular structures by designing the dynamic collective motions of proteins. This concept can lead to the engineering of biocompatible sheet materials, targeted drug transports, and even protein-based nano-robots," comments Prof. Ueno.
Indeed, such developments might just be around the corner.
Reference
Authors :
Kosuke Kikuchi1, Tatsuya Fukuyama2, Takayuki Uchihashi3,4, Tadaomi Furuta1, Yusuke T. Maeda2, Takafumi Ueno1
Title of original paper :
Protein Needles Designed to Self-assemble through Needle Tip Engineering
Journal :
Small
DOI :
Affiliations :
1 School of Life Science and Technology, Tokyo Institute of Technology
2 Department of Physics, Kyushu University
3 Exploratory Research Center on Life and Living Systems, National Institutes of Natural Sciences
4 Department of Physics, Nagoya University
