In vitro, noninvasive, nanoscale characterization of biological macromolecules such as proteins is central to bioengineering research, though current imaging tools often use ionizing radiation under conditions that are far from the molecule’s native biological environment. Powerful imaging techniques such as fluorescence microscopy can potentially damage biological material, and they often do not provide chemical information.
To resolve this challenge, the researchers combined nano-Fourier transform infrared (nano-FTIR) spectroscopy with graphene-capped liquid cells. The imaging platform could open opportunities in the study of soft materials for sectors that range from biology to plastics processing to energy.
The atomically thin graphene membrane that the researchers used was transparent to IR photons; the team acquired atomic force microscopy (AFM) and IR spectroscopy images at nanometer resolution of biological material in physiological liquid, within a cell enclosed by the membrane. As a result, the cell encased by the single-layer graphene served as both a substrate and as an IR-transparent window.
The researchers placed the graphene layer enclosing the liquid cell over a perforated layer of silicon nitride and gold. IR spectra were taken through the graphene at the perforations, which were 1 µm in diameter. This enabled the researchers to study the self-assembly process of proteins at the graphene-aqueous solution interface. The graphene further served as the membrane separating the solution containing the proteins and calcium ions from the nitrogen-gas environment of the AFM tip, thus eliminating the possibility of sample damage and contamination.
Using AFM to monitor the formation of protein lattices and their structural evolution and by recording the amide I and II IR absorption bands, the method showed the noncovalent interaction between proteins and their response to the environment, including ionic strength and solvation. The researchers tracked the chemical and structural evolution of the proteins during self-assembly with a resolution of 10 to 20 nm on the order of the AFM tip size.
After assembly, the spatial distribution of the amide band intensities revealed inhomogeneities among protein domains not visible in previously reported microscopy results. The work also showed that the evolution of the protein substructure depended on many environmental factors.
The researchers’ approach could be used for operando characterization of proteins and other biological materials. The team anticipates that the platform will provide new molecular-level knowledge about various complex biological systems and processes, including the misfolding of beta amyloid proteins and the voltage-induced evolution of ion-channel proteins.
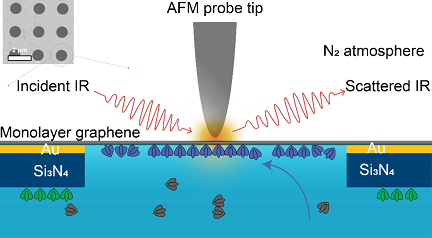
The researcher’s platform comprised AFM topographic imaging at Berkeley Lab’s Molecular Foundry and synchrotron IR nanospectroscopy (SINS) at ALS Beamlines 2.4 and 5.4.
The research was published in PNAS (www.doi.org/10.1073/pnas.2200019119).

