DNA-PAINT is a single-molecule localization microscopy technique that supports multiplexed imaging of fluorescently tagged molecules at high spatial resolution.
Using this approach, the team achieved a localization precision in 3D of less than 2 nm, with axial precision below 0.3 nm. By achieving molecular and even submolecular resolution, 3D superresolution microscopy with nanometer precision will enable new insights into nanostructures and biological systems. The synergetic combination of pMINFLUX and GET allows for superresolution imaging near the surface, for investigating cell adhesion and membrane complexes, for example.
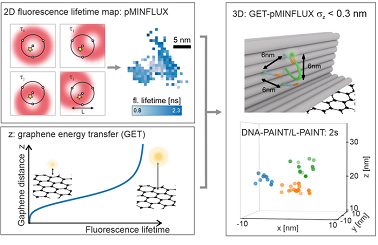
pMINFLUX was introduced by the LMU lab in 2020 as a simplified version of MINFLUX (minimal photon FLUX), a superresolution imaging method that itself combines aspects of single-molecule localization microscopy and stimulated emissions depletion microscopy. In addition to being simpler than the method on which it is built, pMINFLUX also provides the fluorescence lifetime.
MINFLUX interrogates the location of each molecule by focusing a laser near the molecule and measuring the molecule’s distance from the center of the laser focus according to the fluorescence intensity. Researchers can obtain the exact position of the molecule by placing the center of the laser focus at different, successive positions in relation to the molecule.
When pMINFLUX and GET are combined, the information of each photon can be synergistically used for both 2D and axial localization information. Binding sites are placed precisely in 3D by using DNA origami nanopositioners, fluorescent molecules, and DNA-PAINT. The nanopositioners are then used to evaluate the combination of GET, pMINFLUX, and DNA-PAINT for 3D localization and 3D superresolution imaging at different distances from the graphene.
Due to the use of GET, the fluorescence lifetime is distance-dependent; therefore, a change in the fluorescence lifetime can be converted to axial distance from graphene.
This enables GET-pMINFLUX to provide 3D localization with an axial precision of less than 3 Å.
“With the combination of pMINFLUX and graphene, the information of a photon is used in the best possible way,” professor Philip Tinnefeld said. “Here, pMINFLUX is synergistically combined with graphene energy transfer (GET) to enable precisions so far unreached for fluorescence microscopy.”
“The high precisions achievable with GET-pMINFLUX enable the development of methodologically new approaches to improve superresolution microscopy which were previously not realizable,” researcher Jonas Zähringer said. “Typically, the entire photon budget of a dye is needed to localize a position with 1-nm precision. However, GET-pMINFLUX is so photon-efficient that the photon budget can be split among multiple positions and still yield nanometer-precise localizations.”
To overcome the comparatively small field of view of pMINFLUX and the limited binding kinetics of DNA-PAINT, the researchers developed local PAINT (L-PAINT) using DNA nanotechnology. In contrast to DNA-PAINT, which enables superresolution by binding and unbinding a DNA strand labeled with fluorescent dye, L-PAINT has two binding sequences. The researchers designed a binding hierarchy in which the L-PAINT DNA strand binds for longer times on one side. This allows the other end of the strand to scan for binding sites with a high local concentration. The researchers demonstrated L-PAINT by imaging a triangular structure with 6-nm side lengths within seconds.
“The combination with GET-pMINFLUX and L-PAINT allows us to study structures and dynamics at the molecular level, which are fundamental for our understanding of cellular biomolecular reactions,” Tinnefeld said.
In the future, GET-pMINFLUX nanoscopy could be used to investigate artificial bilayers, cellular membranes, and adhesion complexes, as well as macromolecular complexes, with nanometer-scale, 3D precision.
The research was published in Light: Science & Applications (www.doi.org/10.1038/s41377-023-01111-8).

